EDIT: my nistats pull request related to this blog post was merged!
Despite a desire to participate in developing open-source software and develop good open science practices, my PhD has involved a lot of proprietary and closed datasets and software due to who I was working with. This summer I have been returning to some fMRI data that I collected ages ago but haven’t gotten around to analyzing due to other projects. Here I want to summarize my experience trying to adopt some better open-science practices and using new tools and standards. I’m coming to this as a relatively seasoned python programmer and data analyzer but having never used these specific tools. I hope this could be useful for someone starting from scratch or someone interested in a newcomer’s experience with these tools.
I did this on my local desktop running Debian 9.9.
Main components:
- Brain Imaging Data Structure (BIDS)
- Datalad
- fmriprep
- nistats/nilearn
Also mentioned:
- Freesurfer
- dcm2niix
- jo
- jq
- PsychoPy
- mriqc
- nipype
- Docker
Step 1: Create your conda environment(s)
I ended up needing to setup two environments, one for BIDS/fmriprep and one for nilearn/nistats due to some conflicting version requirements.
Step 2: Rename Dicoms
Every fMRI analysis package has their own version of this tool which goes through the dicom files as retrieved from the scanner and organizes them into folders with one folder for each acquisition (e.g. T1w, run1). I used freesurfer’s dicom-rename.
Step 3: Create BIDS formatted Datalad dataset
I wanted to use Datalad to version control any changes that I make to the data and also because I ultimately want to share this dataset. I followed the tutorial here which links to a bash script to convert dicoms to nifti files in BIDS data structure format. In retrospect, I should have followed this one which would have made my life easier. My bash scripts here.
Step 3.1: Install Datalad
Step 3.2: Install Dependencies
Both of the tutorials listed above ultimately use dcm2niix to convert dicoms to Nifti files.
- jo
git clone git://github.com/jpmens/jo.git cd jo autoreconf -i ./configure make check make install - jq
sudo apt-get install jq - dcm2niix
conda install -c conda-forge dcm2niix
Step 3.3: Create Nifti files and associated metadata (.json) files
Like I said above, I did this with a [bash script](https://github.com/thompsonj/quilts/blob/master/fmri_data/toNifti.sh but wish I had done it in python with HeuDiConv.
Step 3.4: Create Event files and associated metadata (.json) files
I did this with a bash script that processed the logfiles saved during my experiment which I ran with PsychoPy.
Step 3.5: Verify BIDS validity and add additional info
BIDS specifies how to store information about your participants, tasks and scans in .tsv and .json files.
I used the online BIDS validator to verify everything was named and organized correctly. The validator found a number of errors that I had to correct which helped me to better understand the BIDS format.
Step 3.6: Put the BIDS data into Datalad datasets
If I had used HeuDiConv, I think this could have happened automatically. Instead, I created my Datalad dataset as I converted the dicoms. I honestly didn’t find the Datalad documentation very clear on what the conventions are for organizing a large Dataset, but gathered somewhat indirectly that the idea is to have several nested Datalad datasets. For example, the highest level folder containing your BIDS dataset would be the highest level Datalad dataset, within which each subject folder ‘sub-
Create nested Datalad datasets with datalad create
For example:
cd speechQuiltsfMRI
datalad create -d . sub-1
datalad create -d . sub-2
datalad create -d . sub-3
datalad create -d sub-1 ses-1
datalad create -d sub-1 ses-2
datalad create -d sub-2 ses-1
datalad create -d sub-2 ses-2
...
My dataset structure looks like this:
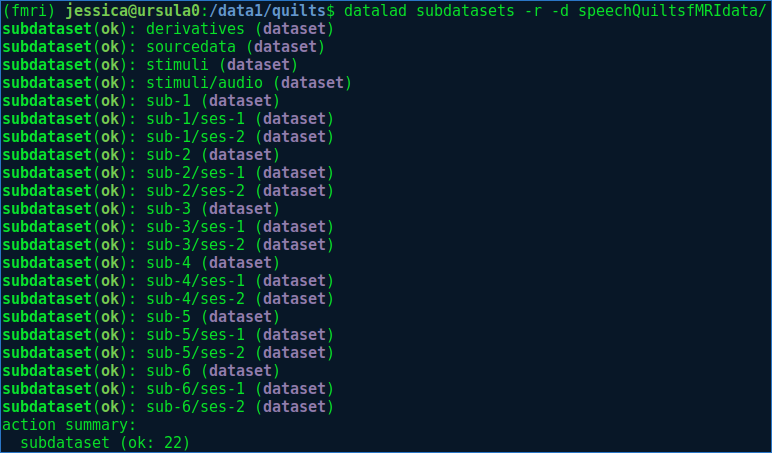
I used datalad add to add directories/files to a dataset. I just noticed that the Datalad documentation says: “Note: This is an obsolete interface. Use datalad.api.save or Dataset.save instead.”, but I don’t see how I would be able to do create these datasets without datalad add.
Save datalad dataset:
datalad save -r -m 'initialize BIDS compliant dataset' .
When all files and directory are added to and saved, running datalad diff -r -d <dataset> should print nothing.
Step 4: Quality Control
At this point I considered using mriqc to perform some quality assurance tests, but for now I skipped this step and went straight on to preprocessing.
Step 5: Preprocessing with fmriprep
fmriprep is built on nipype which “provides a uniform interface to existing neuroimaging software and facilitates interaction between these packages within a single workflow”. fmriprep munges a BIDS dataset, so as long as your data is organized according to BIDS, you can specify your entire preprocessing pipeline with a single call to fmriprep.
Step 5.1: Install Docker and fmriprep-docker
At first I resisted using the Docker container because I am oldschool and like to actually install the software that I’m using. I had many of the dependencies installed already so I thought it would be fine, but quickly ran into runtime issues that I couldn’t easily debug, so I succumbed to using the Docker container.
I followed these instructions for installing Docker including the post-installation steps for Linux which includes how to manage Docker as a non-root user.
Follow this tutorial for the fmriprep docker image.
Aside: I initially missed the line in that tutorial where it says “1) To do the susceptibility distortion correction, the IntendedFor metadata keys need to be defined for every field map that is to be applied.” and since the BIDS specification said that the IndendedFor field was optional, I initially excluded it, assuming that fmriprep would automatically determine that the files in sub-x/ses-y/fmap where intended for all niftis found in sub-x/ses-y/func. That was not the case and in order to get the PE Polar distortion correction to work, I needed to add the IntendedFor field to each .json file in my fmap folders.
Step 5.2: Run fmriprep-docker and save changes the Datalad datasets
I thought that I should be able to use datalad run to call the fmriprep-docker command, which ostensibly should automatically log any changes to the Dataset dataset and the command that led to those changes. So I tried running something like this:
datalad run -d /data1/quilts/speechQuiltsfMRIdata "fmriprep-docker /data1/quilts/speechQuiltsfMRIdata /data1/quilts/speechQuiltsfMRIdata/derivatives participant --participant_label 1 --nthreads 8 --omp-nthreads 16 --mem-mb 64000 -vv -w /data1/quilts/fmriprep_work --resource-monitor --fs-license /usr/local/freesurfer/license.txt"
fmriprep ran but the changes to the Datalad dataset could not be saved automatically because the outputs of fmriprep are saved with limited permissions. I’m not sure I fully understand the philosophy of the restricted permissions…I guess it to prevent users from making unintentional modifications since the idea is that the resulting derivatives should be fixed once created. It sort of backfires in this case where I’m unable to use datalad run. I found this comment on Neurostars which describes how to run the fmriprep docker container under one’s user name instead of root. In my case, I just ran frmiprep-docker directly followed by sudo chown -R <user> <dataset> and datalad save -r -d <dataset> instead.
I experienced some confusion related to how to organize the reference images that I collected for distortion correction. I originally thought that since they were collected with the same scanner protocol as the rest of my runs, that they should have the ‘bold’ suffix. I later realized that they should have the ‘epi’ suffix. Reading the BIDS specification section on fieldmap data I was also perplexed by the reference to Spin Echo EPI scans since, as far as I know, PEPOLAR distortion correction functions like TOPUP also work with Gradient Echo EPI scans. Why the specification of Spin Echo?
I inspected the reports generated by fmriprep to verify what was performed. I love these reports I just wish they were easier to share. I tried to send the html and corresponding images to my advisor but it seemed like the paths to the images were absolute rather than relative.
Step 6: First-level GLM model in nistats
Once the preprocessing was complete, my next goal was to do a simple sanity check to make sure that we see activation in auditory cortex for my auditory stimuli. I decided to use nistats which ostensibly should be able to automatically specify a GLM from preprocesed files in a BIDS dataset. Unfortunately, it didn’t work out of the box on the derivatives saved by fmriprep.
Issues
-
nistats uses different BIDS conventions than fmriprep
Nistats was looking for files named according to older conventions than what was output by fmriprep. This problem was mentioned in an open issue on github.
I created a fork of nistats and updated the relevant functions involved in creating a first level model from a BIDS dataset to use the latest conventions. I eventually submitted these changes as a pull request but it is still in progress. Eventually, it would make sense to do a more major overhaul of nistats to use pybids for this kind of thing.
Note: The current official BIDS specification doesn’t cover derivatives other than to say that derivatives should be in a folder called ‘derivatives’ and should not share file names with their corresponding raw/unprocessed files. However, there is a BIDS Extension Proposal to cover derivatives which has been evolving gradually.
-
fmriprep Confounds fmriprep automatically prepares a number of nuisance variables and saves them in a ‘
desc-confounds_regressors.tsv' file. Some of these confound variables are derivatives (calculus) and so are one element shorter than their counterparts, resulting in a leading NaN for these variables. This led to errors when nistats tried to automatically put these confounds into a design matrix. For now I just omitted these confound variables. One could easily omit only the confound variables including NaNs. Probably a better solution would be to replace the leading NaN with the mean of the variable. -
Missing events in some runs In my experiment, there are 0–3 button presses in every run. Because some runs have no button presses, the design matrix constructed for those runs had one fewer column than the others. This made it impossible to calculate any contrasts. I tried adding a column of zeros to the design matrices for runs without button presses, but this also led to issues. I didn’t finish debugging this problem but it seemed related to this open issue on github. In my case I simply omitted the button presses from the events files since I didn’t care so much about them. This analysis is only a sanity check after all.
With these modifications, I was able to fit first level models and compute basic contrasts like the ones shown below. It looks like my subjects heard the sounds I played for them!
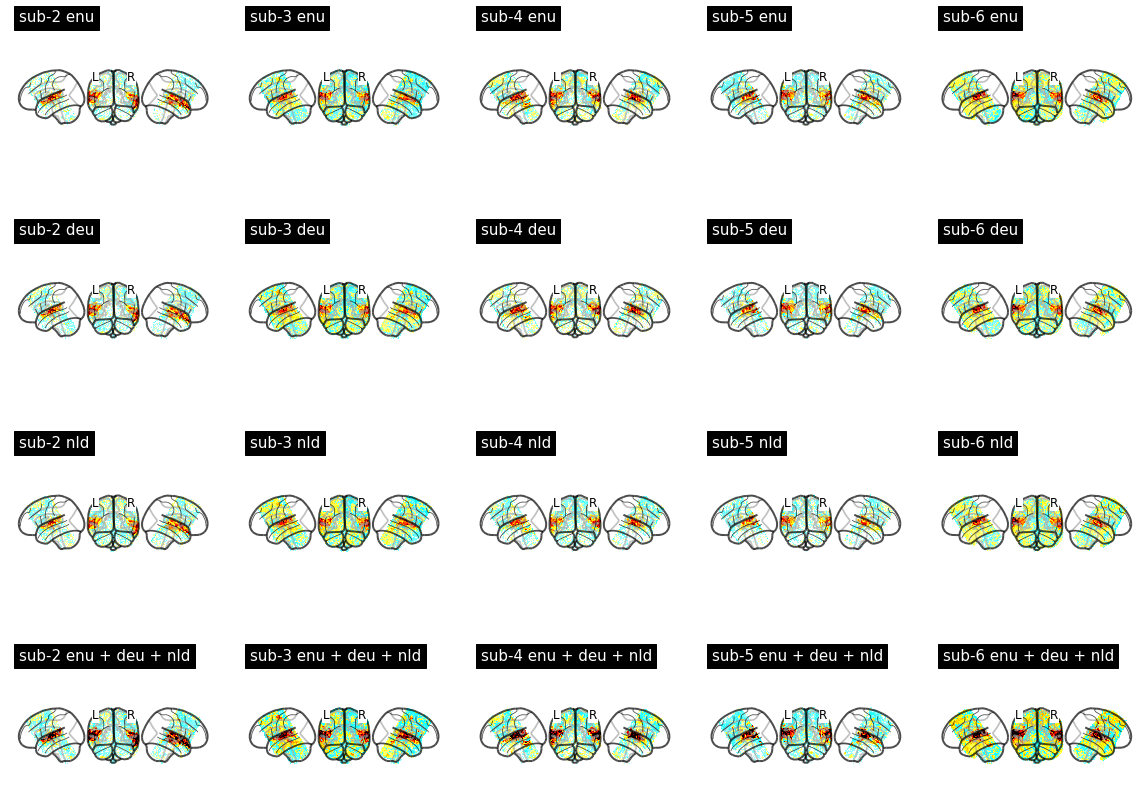
Summary of issues encountered
- Couldn’t use
datalad runto automatically track the effect of running fmriprep due to permissions issues - Confusion about how to organize my epis in opposite phase encoding directions to enable Phase Encoding POLARity (PEPOLAR) susceptibility distortion correction in fmriprep.
- Difficulty sharing the reports generated by fmriprep
- nistats uses different BIDS conventions than fmriprep
- nistats doesn’t deal with NaNs in the confounds prepared by fmriprep
- nistats doesn’t handle different design matrices for different runs